What's next with microarrays? Three- dimensional arrays

CombiMatrix ups the sensitivity by adding a third dimension
By Laura DeFrancesco
Managing Editor, Bioresearch Online
CombiMatrix, the Seattle-based microarray company, has added a new dimension to microarrays—literally. Using a proprietary porous reaction matrix, CombiMatrix fashions three-dimensional arrays that can increase by several orders of magnitude the surface area for capture probes. What does that get you? Greater sensitivity and ease of detection—a simple instrument based on a CCD camera will suffice; no fancy confocal imaging is required.
Personalized microarrays
SNPs on chips
Creating arrays on the Web
About CombiMatrix
Personalized microarrays
But that's not the only thing that sets CombiMatrix apart. Rather than spotting or photolithography, the two most commonly used methods for creating microarrays, the company uses electrochemistry to direct the synthesis of DNA, RNA, peptides, or other biological compounds on pre-fabricated semi-conductor chips. At present, they are working with off-the-shelf chips that have 1,000 sites per square centimeter, but according to Sia Ghazvini, vice president for business development at CombiMatrix, the technology can be down-scaled to 10,000, 100,000, or even a million sites per square centimeter. This union of electrochemistry with semiconductor technology has created a platform that, within a day, can easily create customized microarrays for specific research needs.
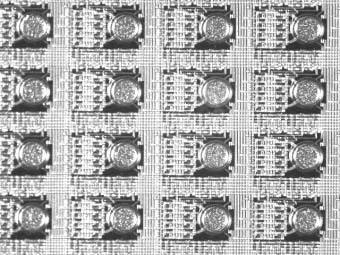
Close-up view of a portion of the array showing round electrodes. DNA synthesis occurs only over electrodes that are turned "on" for that synthesis cycle.
CombiMatrix' microarray chip consists of thousands of tiny chemical "virtual flasks" arranged in a grid pattern on the surface of a semiconductor wafer. The virtual flasks, which are the thickness of a human hair, are separated using carefully engineered liquid solutions rather than physical walls. Each virtual flask has electronic circuitry that is directed by a computer to construct a unique chemical compound, making a system easily customizable to meet the needs of individual researchers.
SNPs on chips
Because of the increased density of capture probe on CombiMatrix chips, signal intensities are achieved that can be several orders of magnitude greater than those obtained from conventional glass or silicon chips. This means hybridization experiments can be performed at higher stringency, enabling the detection of single base-pair mismatches between short sequences.
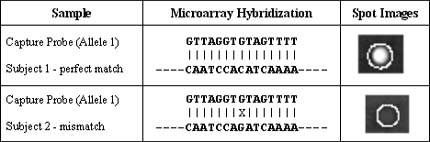
In the example shown above, capture probes corresponding to one allele of a known SNP were synthesized on CombiMatrix microarrays. Samples from two individuals were analyzed for their ability to interact with the capture probe. Each individual's DNA sequence, indicated on the bottom line of each row, was verified independently.
In addition to SNP screening, the signal intensities obtained with CombiMatrix chips allows for SNP discovery. The figure below shows an example of SNP discovery using a CombiMatrix chip. Based on the sequence of a single individual's mitochondrial DNA obtained from GenBank, CombiMatrix designed an array that consisted of 15-mers covering an 800 basepair region, each overlapping by 10 base pairs (a total of 160 spots). Mitochondrial DNA from four ethnically diverse individuals was labeled with either Texas red or fluorescein, hybridized to the chip, and the intensity of the resulting spots was measured.
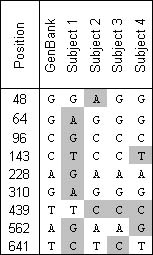 The hypothesis was that strong spots would indicate a perfect match, and that weak signals could indicate a possible mismatch or SNP. Independent sequencing of the mitochondrial DNA from each test sample verified that in every case of weak hybridization, there was a SNP present, and conversely, that the assay was able to detect every instance of a mismatch.
The hypothesis was that strong spots would indicate a perfect match, and that weak signals could indicate a possible mismatch or SNP. Independent sequencing of the mitochondrial DNA from each test sample verified that in every case of weak hybridization, there was a SNP present, and conversely, that the assay was able to detect every instance of a mismatch.
This experiment demonstrates several principles: the ability to detect SNPs by hybridization on a chip surface, the ability to narrow down the location of potential SNPs, the sensitivity of the assay in detecting SNPs that had not previously been identified, and the ability to carry out double-labeling experiments.
Creating arrays on the Web
Now in beta testing, CombiMatrix' microarrays will be ready for the marketplace next year. And along with arrays, the company will be providing a set of bioinformatics tools on its website that will enable users to design and order customized chips from the convenience of their computer.
Internet-enabled bioinformatics software will aid users at both ends of the process. The customer can use the software to optimize capture probe sequences as well as to design the layout of the array. Once the layout has been finalized, the customer can send the order directly to the company's manufacturing equipment via the Internet, accessing a human customer-service intermediary only when needed. Once the customized chip has been obtained and used, the user can use CombiMatrix' software to analyze and interpret the experimental results.
About CombiMatrix
Founded in 1995, CombiMatrix develops research tools for the pharmaceutical and biotechnology industries. Together with its novel biochip array processor system, CombiMatrix is poised to leverage the power of bioinformatics and the speed of the Internet to transform the process of generating, analyzing, and managing complex biological information. This platform technology integrates electrochemical and semiconductor technologies to enable biological applications such as genomics and proteomics. CombiMatrix is located outside of Seattle and is majority-owned by Acacia Research (Pasadena, CA), a developer and operator of technology and Internet companies.
For more information: CombiMatrix, 34935 SE Douglas Avenue, Snoqualmie, WA 98065. Tel: 425-396-5940. Email: info@combimatrix.com.
